降维_线性判别分析
2020.01.16
Lixin
热度
℃
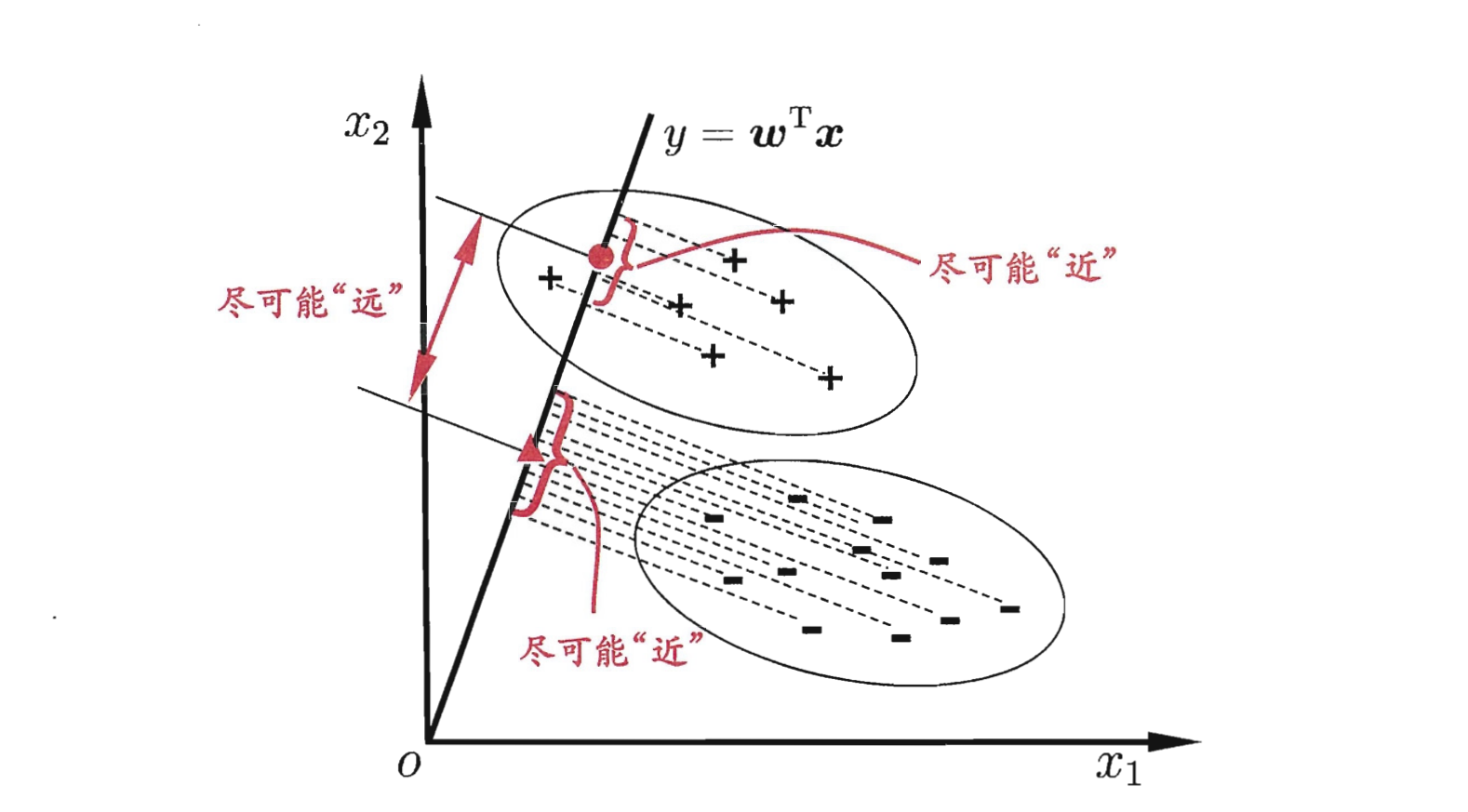
1. 算法概述 LDA的思想可以用一句话概括,就是“投影后类内方差最小,类间方差最大”,如下图所示。我们要将数据在低维度上进行投影,投影后希望每一种类别数据的投影点尽可能的接近,而不同类别的数据的类别中心之间的距离尽可能的大。
2. 算法推导 LDA多分类:假定存在$N$个类,且第$i$类示例树为$m_i$,我们定义全局散度矩阵:
其中$\mu$是所有示例的均值向量。将类内散度矩阵$S_w$定义为每个类别的散度矩阵之和,即
其中
由式$(1)-(3)$可得:
多分类LDA有多种实现方式:使用$S_b,S_w,S_t$中的任意两即可。常见的一种实现是采用优化目标
其中$W \in R^{d\times(N-1)}$,$tr(.)$表示矩阵的迹。式$(5)$可以通过求解如下式的广义特征问题
$W$的闭式解则是$S_w^{-1}S_b$的N-1个广义特征值所对应的特征向量组成的矩阵。
若将$W$视为一个投影矩阵,则多分类LDA将样本矩阵投影到$N-1$维空间。$N-1$通常原小于数据原来的维数,且投影过程中使用了类别信息,因此LDA也被视为一种数据降维的技术。
3. 实现步骤
对于每一类别,计算$d$维数据的均值向量;
构造类间散度矩阵$S_b$和类内散度矩阵$S_w$;
计算矩阵$S_w^{-1}S_b$的特征值及对应的特征向量;
选取前$k$特征值所对应的特征向量,构造$d \times k$维的转换矩阵$W$,其中特征值以列的形式排列;
使用转换矩阵$W$将样本映射到新的特征子空间上。
4. LDA与PCA LDA和PCA都可以用作降维技术,下面比较一下相同点和不同点:
4.1 相同点 1)两者均可以对数据进行降维;
2)两者在降维时均使用了矩阵特征分解的思想;
3)两者都假设数据符合高斯分布.
4.2 不同点 1) LDA是有监督的降维技术,PCA是无监督的降维技术;
2) LDA还可以用于分类,后续在分类时,贴上分类的处理方法;
3) LDA最多可降低到$k$维($k$是分类的个数),而PCA最多可降低到$n-1$维($n$是数据的维数);
4) LDA选择分类性能最好的投影方向,而PCA选择样本点投影具有最大方差的方向。
5. LDA的实现代码 5.1 Scala lda实现 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 package ml.scrath.ldaimport breeze.linalg._import breeze.stats._import org.apache.spark.rdd.RDD class LinearDiscriminantAnalysis extends Serializable def fit RDD [DenseVector [Double ]], labels: RDD [Int ],k:Int ) = { val sample = labels.zip(data) computeLDA(sample,k) } def computeLDA RDD [(Int , DenseVector [Double ])],k:Int )= { val featuresByClass = dataAndLabels.groupBy(_._1).values.map(x => rowsToMatrix(x.map(_._2))) val meanByClass = featuresByClass.map(f => mean(f(::, *))) val Sw = featuresByClass.zip(meanByClass).map(f => { val featuresMinusMean: DenseMatrix [Double ] = f._1(*, ::) - f._2.t featuresMinusMean.t * featuresMinusMean: DenseMatrix [Double ] }).reduce(_+_) val numByClass = featuresByClass.map(_.rows : Double ) val features = rddToMatrix(dataAndLabels.map(_._2)) val totalMean = mean(features(::, *)) val Sb = meanByClass.zip(numByClass).map { case (classMean, classNum) => { val m = classMean - totalMean (m.t * m : DenseMatrix [Double ]) :* classNum : DenseMatrix [Double ] } }.reduce(_+_) val eigen = eig((inv(Sw ): DenseMatrix [Double ]) * Sb ) val eigenvectors = (0 until eigen.eigenvectors.cols).map(eigen.eigenvectors(::, _).toDenseMatrix.t) val topEigenvectors = eigenvectors.zip(eigen.eigenvalues.toArray).sortBy(x => -scala.math.abs(x._2)).map(_._1).take(k) val W = DenseMatrix .horzcat(topEigenvectors:_*) (W ,Sb ,Sw ) } def rowsToMatrix TraversableOnce [DenseVector [Double ]]): DenseMatrix [Double ] = { rowsToMatrix(in.toArray) } def rowsToMatrix Array [DenseVector [Double ]]): DenseMatrix [Double ] = { val nRows = inArr.length val nCols = inArr(0 ).length val outArr = new Array [Double ](nRows * nCols) var i = 0 while (i < nRows) { var j = 0 val row = inArr(i) while (j < nCols) { outArr(i + nRows * j) = row(j) j = j + 1 } i = i + 1 } val outMat = new DenseMatrix [Double ](nRows, nCols, outArr) outMat } def rddToMatrix RDD [DenseVector [Double ]]): DenseMatrix [Double ] = { val inArr = inArr1.collect() val nRows = inArr.length val nCols = inArr(0 ).length val outArr = new Array [Double ](nRows * nCols) var i = 0 while (i < nRows) { var j = 0 val row = inArr(i) while (j < nCols) { outArr(i + nRows * j) = row(j) j = j + 1 } i = i + 1 } val outMat = new DenseMatrix [Double ](nRows, nCols, outArr) outMat } }
5.2 scala 测试代码 测试代码(其中数据iris.csv是由下面python代码生成)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 package ml.scrath.ldaimport org.apache.spark.sql.SparkSession import breeze.linalg.DenseVector object TestLDA extends App val spark = SparkSession .builder() .appName("DataFrame-Basic" ) .master("local[4]" ) .config("spark.sql.warehouse.dir" , "file:///E:/spark-warehouse" ) .getOrCreate() val sc = spark.sparkContext val irisData = sc.textFile("D:\\data\\iris.csv" ) val trainData = irisData.map { _.split("," ).dropRight(1 ).map(_.toDouble) }.map(new DenseVector (_)) val labels = irisData.map { _.split("," ).apply(4 ).map(_.toInt).apply(0 ) } val start = System .currentTimeMillis() val model = new LinearDiscriminantAnalysis val k = 2 val res = model.fit(trainData, labels, k) println("=====W======" ) println(res._1) println("=======Sb====" ) println(res._2) println("=======Sw====" ) println(res._3) }
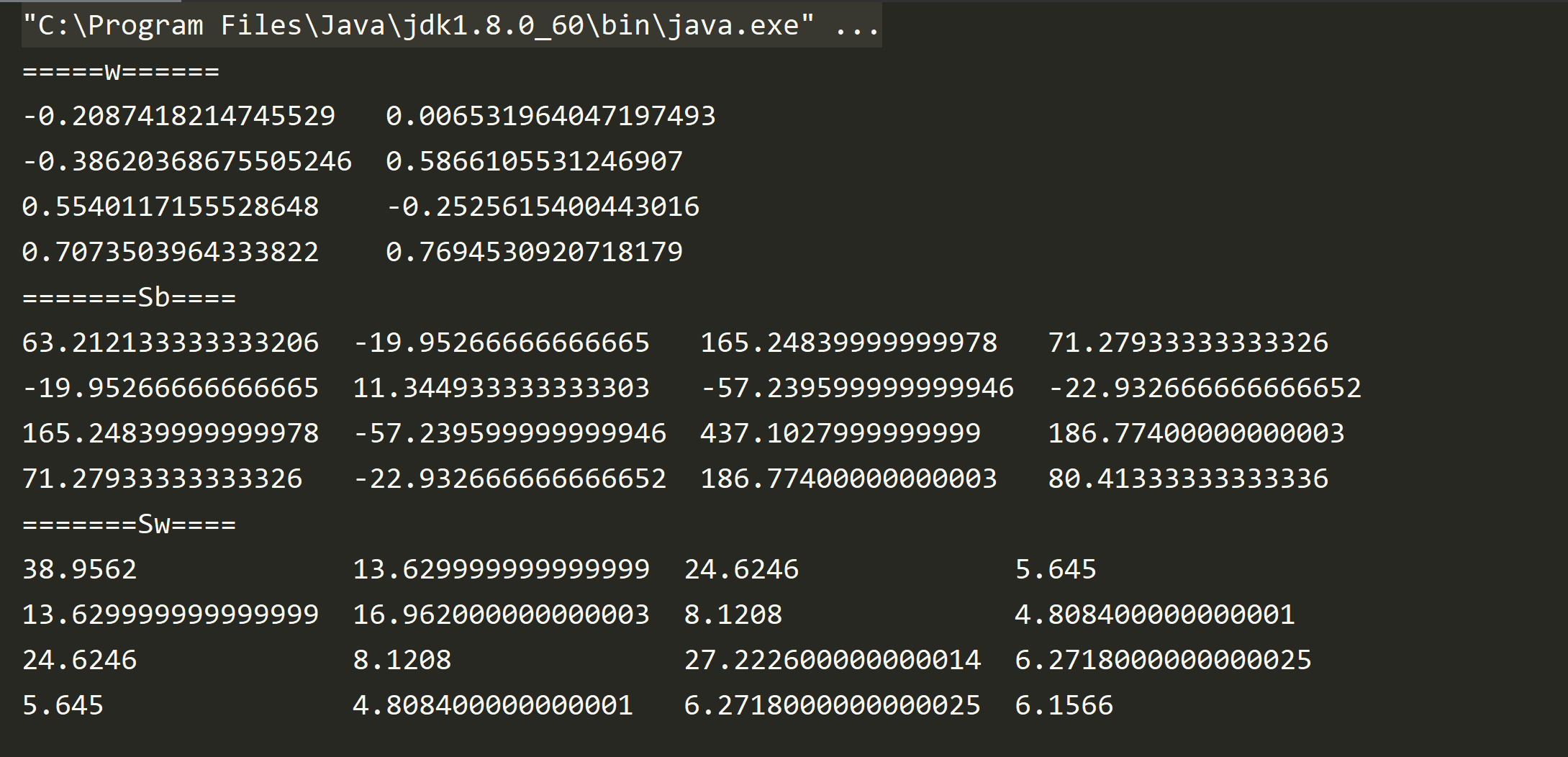
5.3 scala 结果 展示转换矩阵$W$, 类间散度矩阵$S_b$和类内散度矩阵$S_w$
5.4 Python lda 代码 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 import numpy as npfrom sklearn.datasets import load_irisimport matplotlib.pyplot as pltfrom sklearn.discriminant_analysis import LinearDiscriminantAnalysisdef LDA (X, y, k) : ''' X为数据集,y为label,k为目标维数 ''' label_ = np.unique(y) mu = np.mean(X, axis=0 ) Sw = np.zeros((len(mu), len(mu))) for i in label_: _X = X[y == i] _mean = np.mean(_X, axis=0 ) Sw += np.dot((_X - _mean).T, _X - _mean) print(Sw) Sb = np.zeros((len(mu), len(mu))) for i in label_: _X = X[y == i] _mean = np.mean(_X, axis=0 ) Sb += len(_X) * np.dot(( _mean - mu).reshape( (len(mu), 1 )), (_mean - mu).reshape((1 , len(mu)))) print(Sb) eig_vals, eig_vecs = np.linalg.eig(np.linalg.inv(Sw).dot(Sb)) sorted_indices = np.argsort(eig_vals) topk_eig_vecs = eig_vecs[:, sorted_indices[:-k - 1 :-1 ]] return topk_eig_vecs,Sb,Sw if '__main__' == __name__: iris = load_iris() X = iris.data y = iris.target data_cat = np.c_[X,y] import pandas iris_df = pandas.DataFrame(data_cat) iris_df.to_csv("iris.csv" ,index = 0 ,header = False ) W,Sb,Sw = LDA(X, y, 2 ) X_new = np.dot((X), W) plt.figure(1 ) plt.scatter(X_new[:, 0 ], X_new[:, 1 ], marker='o' , c=y) print("========W=========" ) print(W) print("========Sb========" ) print(Sb) print("========Sw========" ) print(Sw) lda = LinearDiscriminantAnalysis(n_components=2 ) lda.fit(X, y) X_new = lda.transform(X) plt.figure(2 ) plt.scatter(X_new[:, 0 ], X_new[:, 1 ], marker='o' , c=y) plt.show()
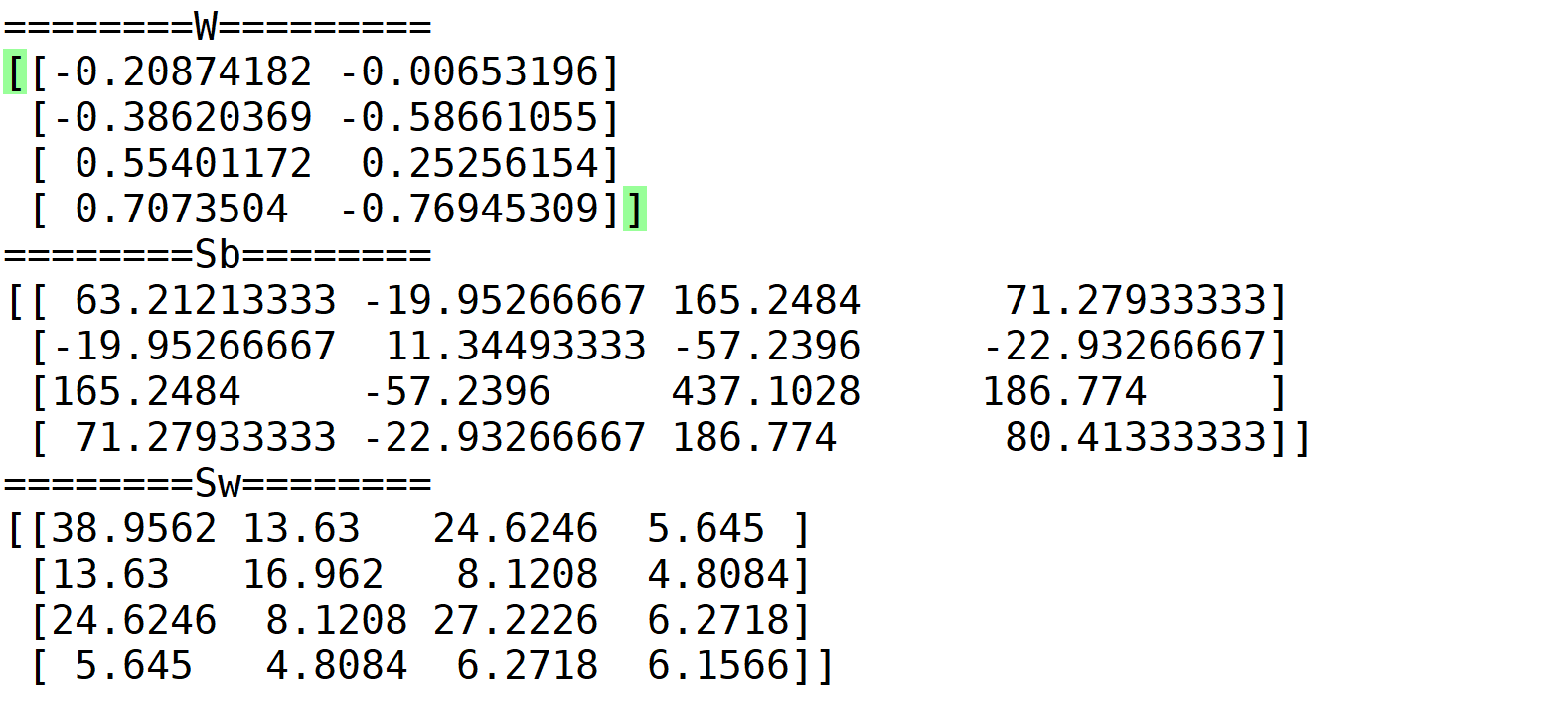
5.5 Python 结果 展示转换矩阵$W$, 类间散度矩阵$S_b$和类内散度矩阵$S_w$,可见scala和python得到结果是一致的。